A brief, preliminary evaluation
For the revision of a new manuscript, I needed to generate a figure, in which a phylogenetic tree is plotted over a geographic map. The tips of the tree shall hereby point to the distribution area of the taxa they represent on the underlying map.
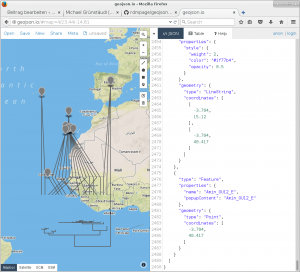
Lucky for me, Rod Page has recently published a small script, which plots the tips of a phylogenetic tree (stored in nexus-format) to latitude and longitude coordinates, which are saved as a geographic character alongside the tree.
php make_html.php MyTree.tre > MyTree.geojson
Upon running the script, I loaded the resulting geojson-file into the online tool geojson.io in order to evaluate the validity of the above-mentioned script. From the looks of it, this script does what it is supposed to do.
In order to adjust line colour and thickness, I loaded the geojson-file into R using the rgdal library and plotted the results.
library(rgdal)
points = readOGR(dsn = "MyTree.geojson", layer = "OGRGeoJSON", require_geomType=c("wkbPoint"))
lines = readOGR(dsn = "MyTree.geojson", layer = "OGRGeoJSON", require_geomType=c("wkbLineString"))
plot(lines)
plot(points, add=T)
In summary, my first impression of this mapping procedure is quite positive.


 Die
Die
Am 12. November 2015 um 13:42 Uhr
Nice post. I know the Robinson and Winkel projections work betetr overall, but being a Kiwi I prefer the old fashioned Mercator projection if only because the others tend to turn New Zealand into a disortorted smear at the edge of the world